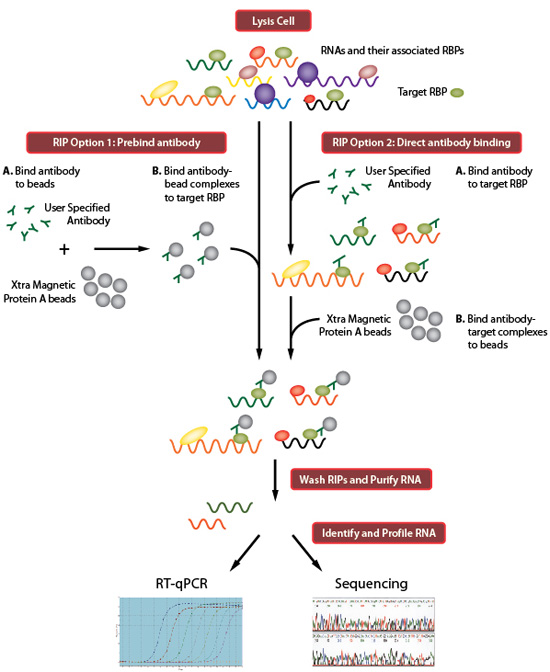
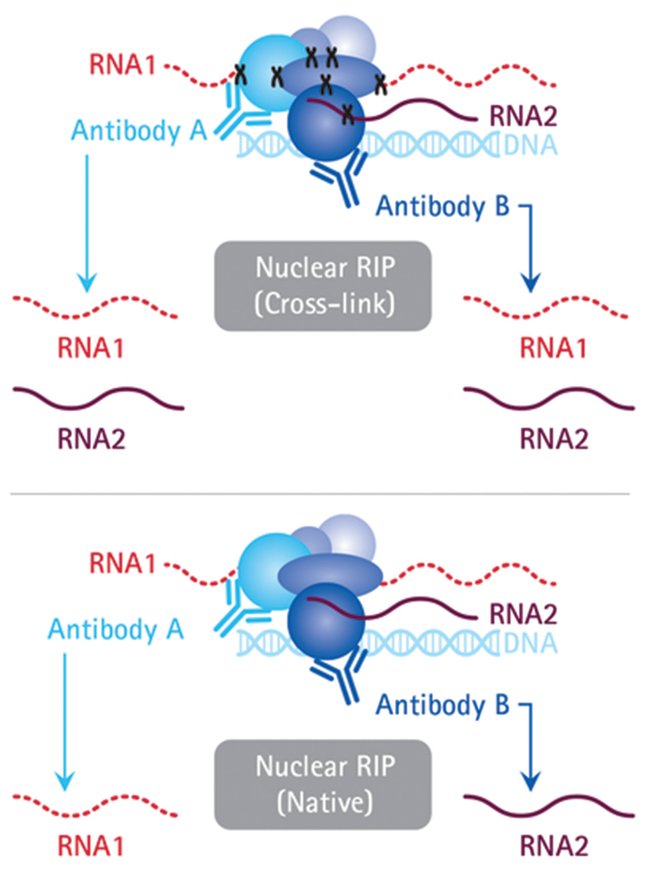
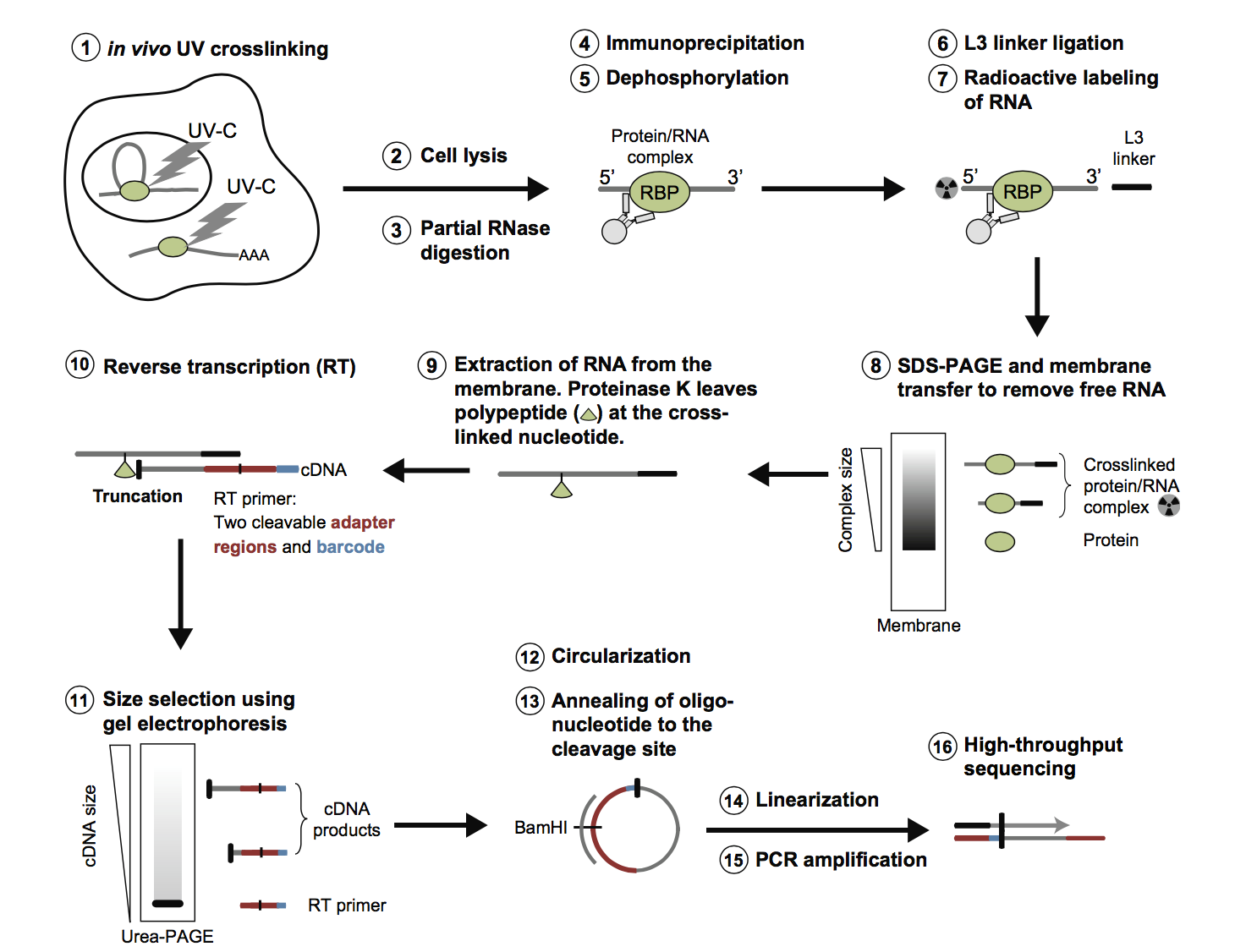
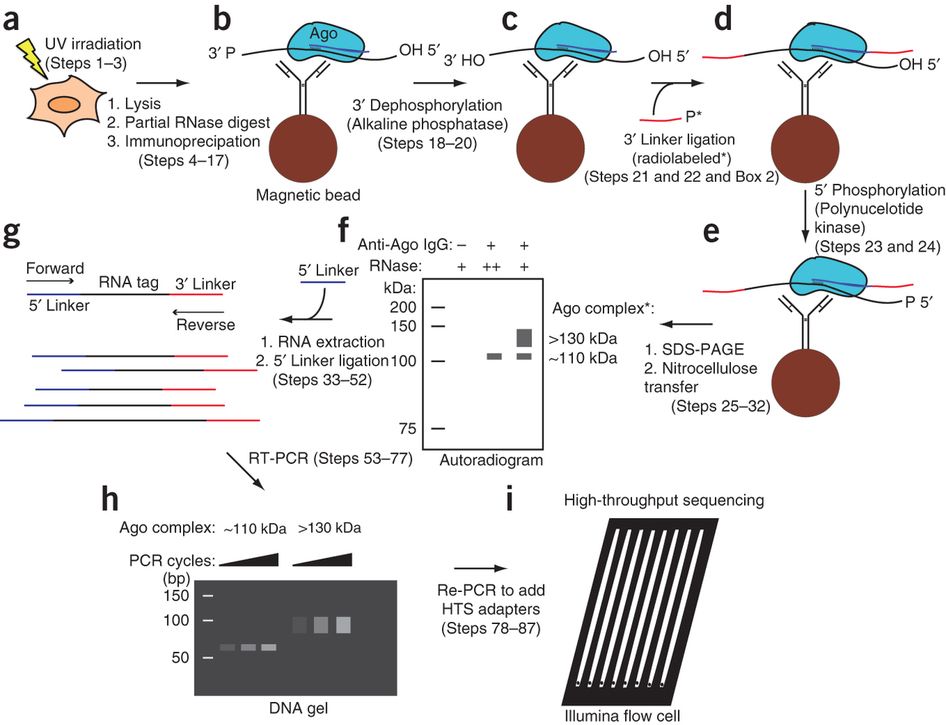
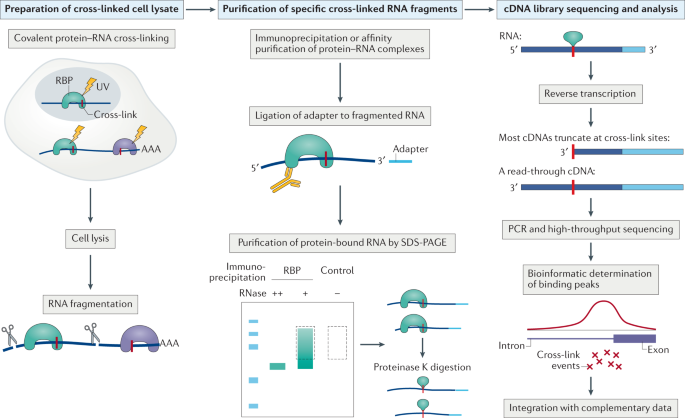
ACE-score-based Analysis of Temporal miRNA Targetomes During Human Cytomegalovirus Infection Using AGO-CLIP-seq

Pipeline for extracting MIWI CLIP-Seq features (see Section 2). The RNA... | Download Scientific Diagram
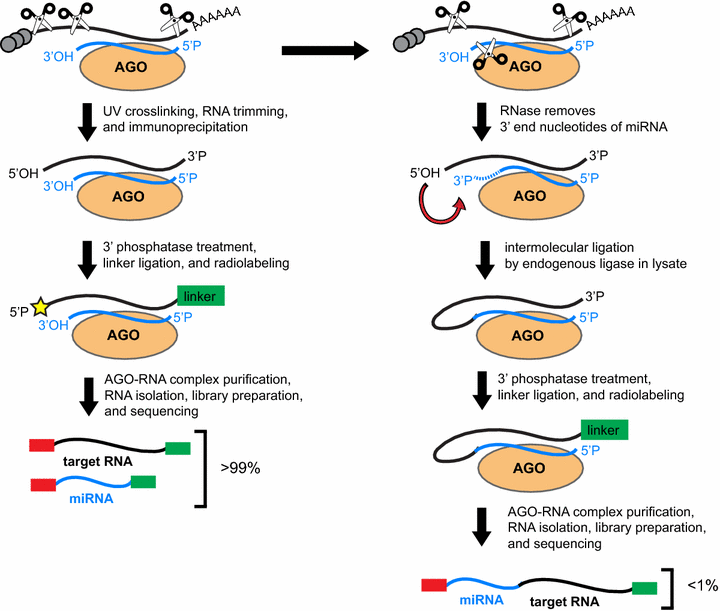
MPs | Free Full-Text | Studying miRNA–mRNA Interactions: An Optimized CLIP-Protocol for Endogenous Ago2-Protein
Analysis of the nucleocytoplasmic shuttling RNA-binding protein HNRNPU using optimized HITS-CLIP method | PLOS ONE

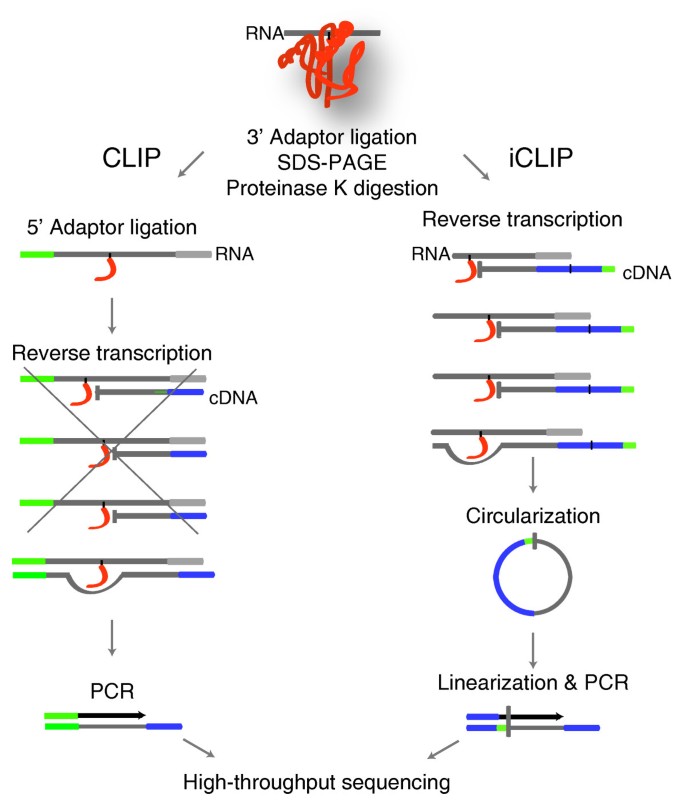
Schematic representation of the iCLIP protocol. Protein-RNA complexes... | Download Scientific Diagram
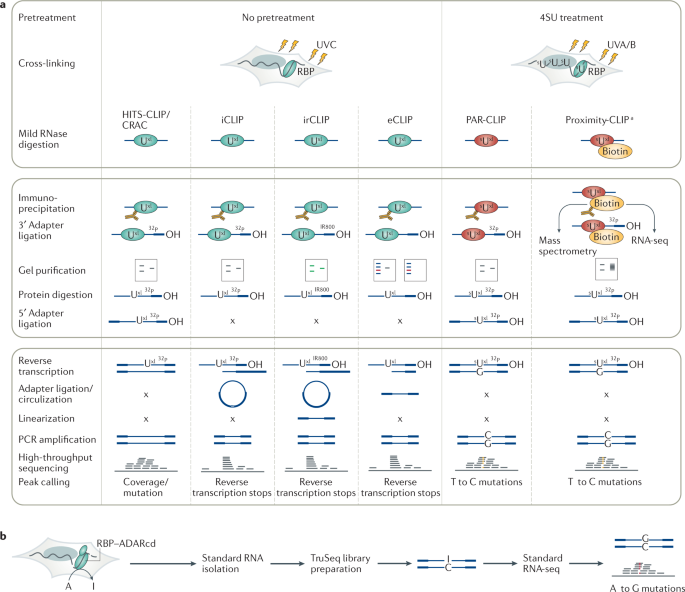
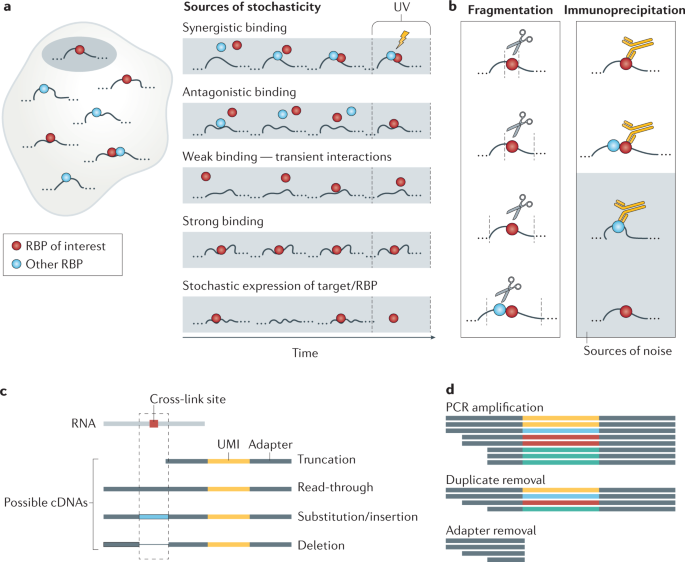



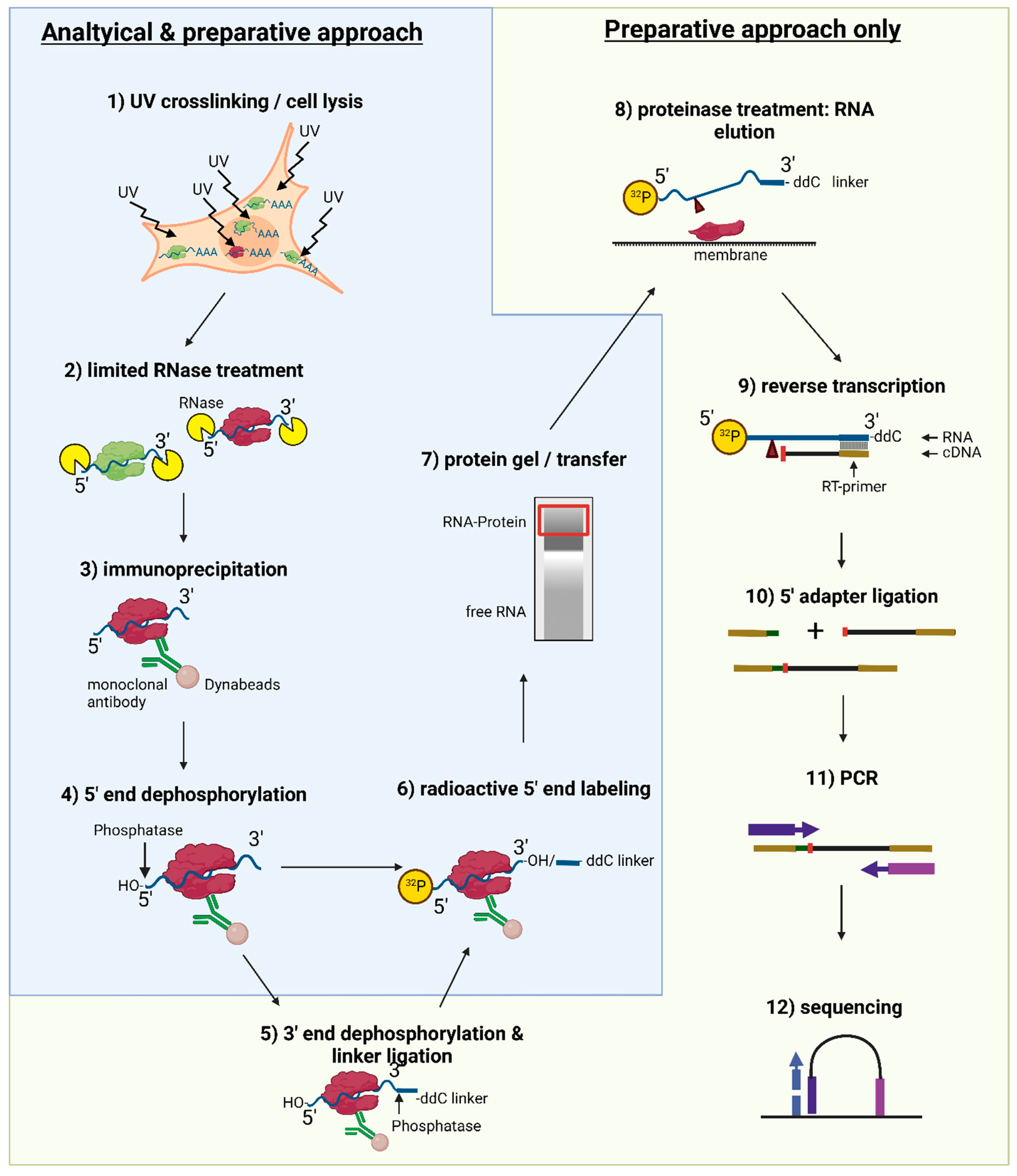
![PDF] CLIPSeqTools—a novel bioinformatics CLIP-seq analysis suite | Semantic Scholar PDF] CLIPSeqTools—a novel bioinformatics CLIP-seq analysis suite | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/6bbd241680ec0f3d6a86dec738a6b864ea5048c3/2-Figure1-1.png)
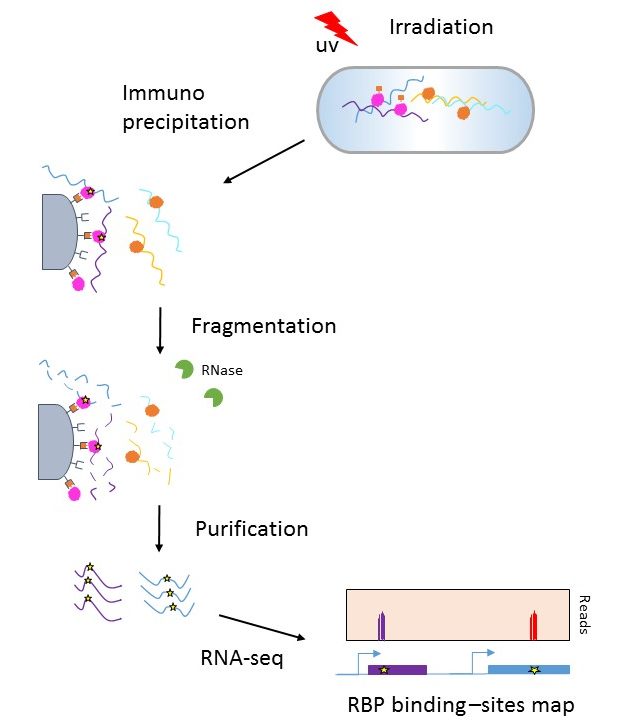
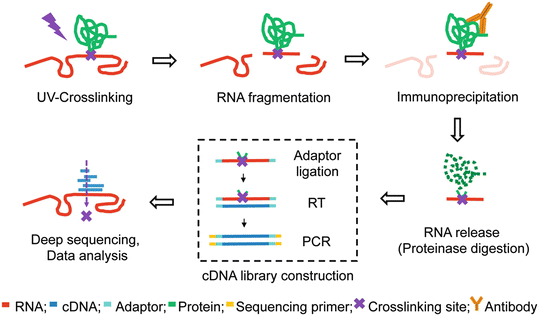

-2.jpg)